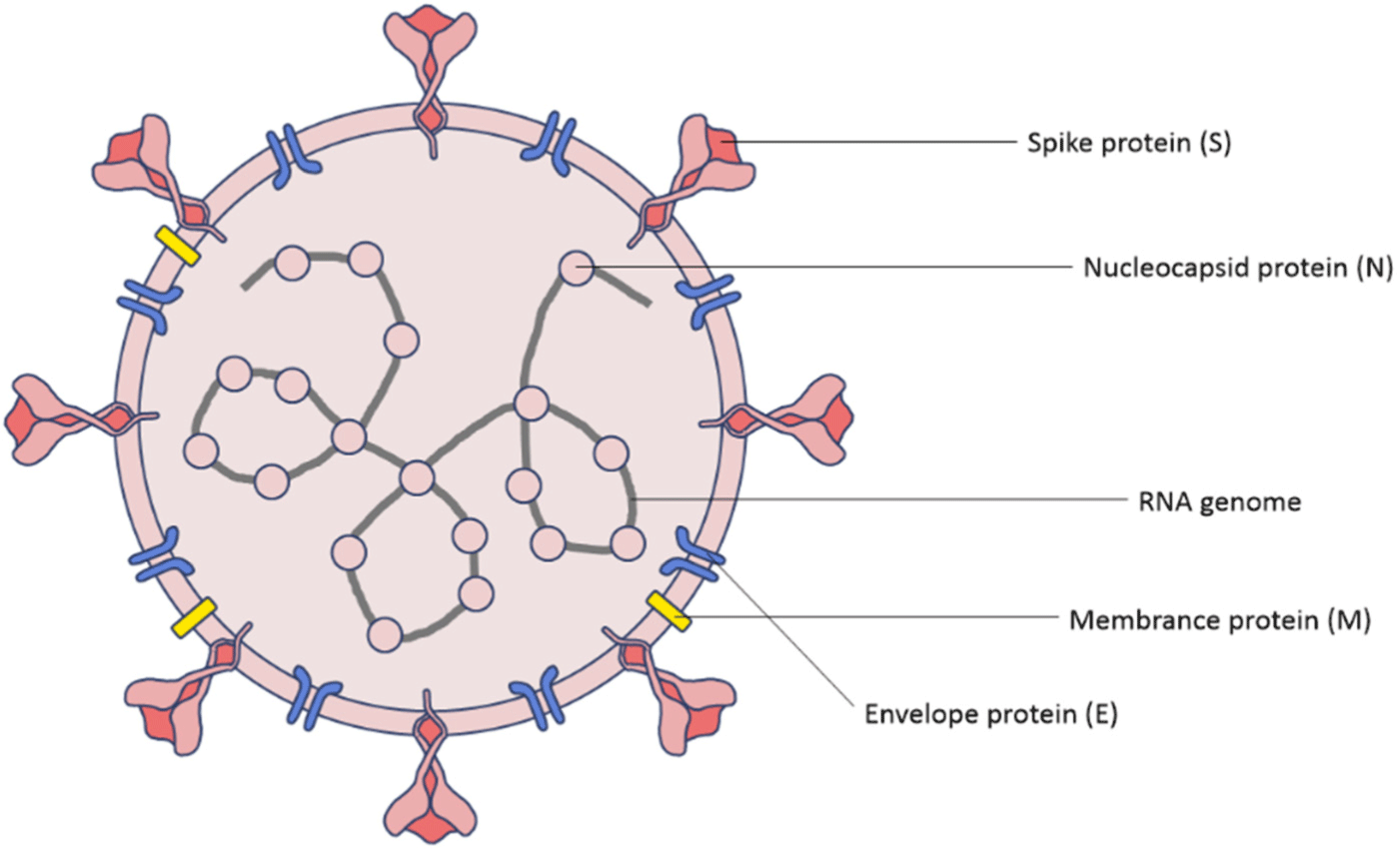
1. The strain of COVID-19
By 25th October 2022, there have been 625,248,843 confirmed cases of coronavirus disease 2019 (COVID-19) worldwide, including 6,562,281 of them resulting in death reported to the World Health Organization (WHO). A total of 12,830,378,906 vaccine doses have been administered (Fig. 1).[1]
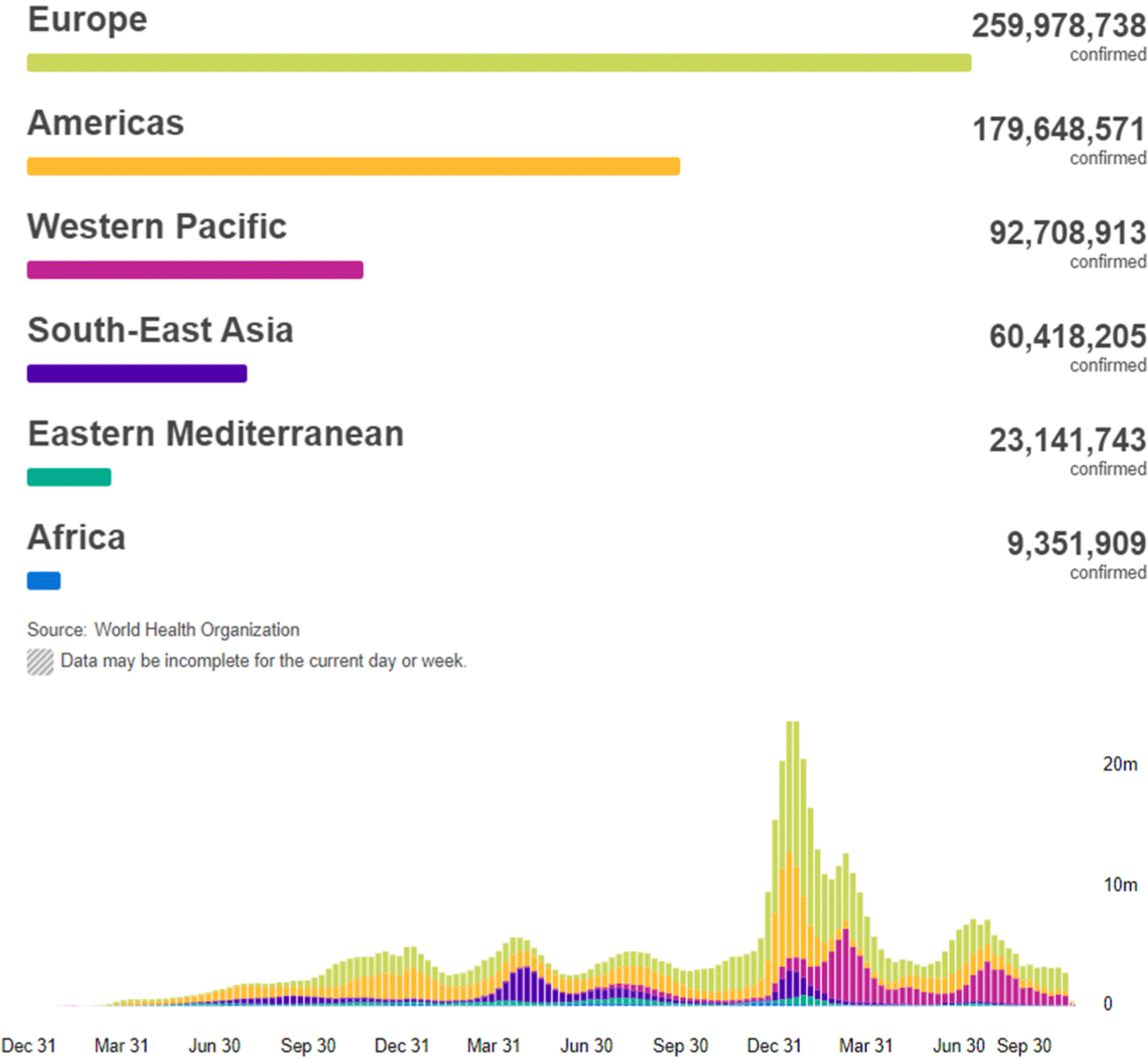
An accurate measurement of the number of deaths due to the COVID-19 pandemic is crucial for the management of the disease in each country and region.[2] Accurate measurement of deaths is also needed to understand the determinants of variation in the infection–fatality ratio across populations, and is a direct input into forecasting the pandemic and investigating alternative health policy options. The WHO reported deaths in an attempt to quantify the magnitude of the COVID-19 pandemic in different populations and locations over time.[3]
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first identified as a respiratory virus by a pneumonia patient in Wuhan, Hubei Province, China in December 2019.[4] As a result of the novel coronavirus gene base analysis, it was confirmed that it had a 89.1% similarity to the bat severe acute respiratory syndrome (SARS)-like coronavirus genome.[5] It is estimated that the SARS-CoV-2, which was parasitic in bats, spread to humans through mutations of wild animals as intermediate hosts. It is estimated that human-to-human transmission is mainly transmitted through respiratory droplets caused by the cough of an infected person.[6] It is mainly accompanied by symptoms such as fever, cough, and difficulty breathing. Fatal cases are accompanied by respiratory failure and severe pneumonia symptoms, and the onset of symptoms is estimated to be between 2 and 14 days after SARS-CoV-2 infection.[7]
To confirm a SARS-CoV-2 case, a swab taken from the nose or throat was examined.[8] Blood tests checked the immune system for antibodies formed in order to defend against SARS-CoV-2.[8] However, time is required for the body to make antibodies, patients were vulnerable the first infection of the disease.[9] After contracting the SARS-CoV-2, patients were required to take acetaminophen or ibuprofen in order to treat the fever and pain.
RNA-based viruses are unstable and mutate easily. In addition, since viruses are parasitic in cells of other life forms, they have the ability to change themselves to suit the nature of the host, and they are very small in size, resulting in poor ability to protect their genes.[10] RNA-based viruses also mixe well with genes and malfunctions very frequently in the process of self-replicating. This mutation is a natural phenomenon that occurs in all living things and is the driving force of evolution. It occurs randomly, and among the mutants, the individual mutant which best adapts to the environment survives.[11] When a virus infects a cell, it creates thousands of self-replications and sometimes errors in the process. These errors are generally harmless and do not cause much change, but sometimes these genetic changes make the virus more contagious and easy to spread. The process of the virus multiplying into host cells and becoming susceptible to human infection is called "mutation", and when such genetic mutations accumulate as human-to-human infections continue to occur, the affinity with human cells increases, which is called a "mutant virus".[11]
As the COVID-19, which occurred in December 2019, spread around the world, it continually mutated into viruses such as alpha, beta, gamma, delta, and omicron viruses.[12] The infection of delta mutant virus originating from India has decreased since April when it spread rapidly between countries and became the dominant species, and then the omicron which was reported in South Africa in November became the dominant species and is currently prevalent around the world.[12] The Korea Disease Control and Prevention Agency is conducting genetic surveillance to track and analyze the source of infection of the SARS-CoV-2 and checking for genetic mutations.[13] As a result of conducting genomic analysis and mutated polymerase chain reaction (PCR) analysis on positive samples of confirmed patients related to domestic infection and overseas inflow, a total of 118,450 major mutated viruses (92,271 delta, 22,703 omicron, 3,296 alpha, 153 beta, and 27 gamma) were identified from 1 January 2020 to 5 February, 2022.[14] In overseas inflow cases, after the delta was first confirmed in Korea in April 2021, delta cases were most confirmed with 39.3% in May and 76.2% in June, but the trend changed with 52.8% of the cases being omicron in December, and 96.4% in January 2022.[14] In the case of domestic infection, the delta mutation became the dominant species with a detection rate of 59.6% in July 2021, but the dominant species in Korea changed into the omicron with a detection rate of 63.9% in January 2022. As a result of analyzing the detailed systems of mutated viruses through electrical genetic analysis, the share of the detailed systems of omicron confirmed in the domestic infection case in January was 3.2% for BA.1.15, 7% for BA.1.1, and 1.4% for BA.2. The Korea Disease Control and Prevention Agency showed that quick detecting and analyzing mutated viruses is essential in preventing the spread of COVID-19, and continues to monitor the emergence of new mutated viruses.[14]
As a result of analyzing the global mutated virus status registered in the COVID-19 gene database, Delta occupied more than 90% from July, but decreased from December with the appearance of Omicron in November, 2021.[15] Omicron, which showed high propagation power, has spread around the world with a 42.1% of the cases in December and 95.1% in January, making it a dominant species worldwide. Sorted by major countries, the omicron dominance was confirmed in November in South Africa, which first reported omicron to the WHO, and also confirmed in December, 2021 in the UK, Australia, the United States, Israel, Denmark, Spain, and Singapore. South Korea, along with New Zealand and Lithuania, became the dominant species in mid-January, 2022.[16] With the diffusion of omicrons, the system of omicrons was subdivided into the BA.1, BA.1.1, BA.2, and BA.3 systems from the existing BA.1.1.529 system. In January, 2022, BA.1 had 54.3%, BA.1.1 had 34.1%, and BA.26.7%, respectively, but the recent increase in BA.2 has been confirmed as the most common case, especially in Denmark, India, South Africa, Nepal, and the Philippines.[16] Unlike BA.1, BA.2 has a characteristic of having no fruits of spike (S) protein, compared to the usual 69 to 70. PCR diagnostic reagents used in some countries, including the UK, were used to estimate omicron cases along with COVID-19 diagnosis, but the BA.2 showed no 69 to 70 results, which made S protein is not being detected during PCR diagnosis (spike gene target file), leading it also to be called the "stealth omicron".[17] Initial limited research results show that BA.2 has higher propagation power than BA.1, and the hospitalization rate and vaccine effect are similar against the BA.1 and BA.2, but further studies are needed to confirm this.[17]
Mutated virus refers to cases in which the transmission power increases or negative changes are confirmed, and when the pathogenicity increases, the change in the severity of the disease is clinically confirmed.[18] It also refers to a mutated virus that has been confirmed to degrade the effectiveness of existing diagnoses, vaccines, and treatments. The omicron-type mutant virus has two to three times higher propagation power than the delta-type mutant, and the possibility of re-infection of the omicron mutant virus is reduced after other mutant infections. In addition, the possibility of hospitalization and severity are reduced and the effects of vaccines are lower. Omicron virus also has a higher infectivity rate of 79.2 percent, but lower mortality rate of 65.0 percent.[19]
The Alpha virus is a mutated virus with a 87% infectivity and 61% fatality rate. The alpha virus first occurred in the UK, with more than 1.27 million cumulative confirmed patients reported worldwide.[19] Beta viruses were 50% more infectious, 57% more lethal, and significantly reduce neutralization by antibodies. The beta virus had about 36,000 cases with the first outbreak happening in South Africa.[20] The Gamma virus also had a high infection rate of 161 percent and a high fatality rate of 50 percent. The Gamma virus infected about 113,000 people with its first outbreak in Brazil. The Epsilon virus is a mutation that is 22% more infectious than original strain. The Epsilon virus was first detected in the United States, with about 75,000 people infected worldwide. The Delta virus is 198% more infectious and 85% more lethal than original strain. The delta virus has infected more than 100 million people worldwide and was the first to occur in India.[20]
2. The deterioration of COVID-19
The incubation period for SARS-CoV-2 infection is 1 to 14 days, and the virus is able to be detected 1 to 3 days before the onset of symptoms. The symptoms of the COVID-19 are similar to other respiratory diseases such as the flu and colds, but the symptoms may vary from person to person, and symptoms may range from asymptomatic to very severe symptoms.[21] The most common symptoms are fever, cough, fatigue, and with others as abnormal smell and taste, muscle pain, sore throat, runny nose, stuffy nose, headache, diarrhea, and skin symptoms.[21] Symptoms usually appear about 2 to 14 days after infection and resolve after about 1 week, but some may persist for several weeks. Most recover without special treatment, but some exacerbate to severe symptoms such as pneumonia or shortness of breath. In addition, as SARS-CoV-2 evolves, new additional mutations appear, and it is known that transmission power or infection pattern is different for each mutation.[22] In the case of the omicron mutation, the infection spreads easily and quickly, but the severity is lower than that of the delta mutation.[23] In terms of the infection pattern, it is known that omicron mutations mainly cause upper respiratory tract infections, whereas delta mutations caused lower respiratory tract infections such as rapid progression of pneumonia (Fig. 2).
A virus in the coronavirus (CoV) family that causes COVID-19 is a variant of SARS-CoV. Unlike conventional coronaviruses, the SARS-CoV-2 has a mutation in the ‘spike’, an external device for contacting respiratory epithelial cells.[24] This spike glycoprotein which forms trimers on the surface of virions is the main determinant of coronavirus tropism.[25] Normally, the immune system focuses on the spikes of viruses which stick to the cells, but the SARS-CoV-2 mutates into spikes that are unfamiliar to the immune system, which results in the immune system difficult to detect it. The spike protein is divided into two subunits, S1 and S2.[24] The S1 binds itself to the host entry receptor angiotensin-converting enzyme 2 (ACE2) and the S2 mediates membrane fusion. This activates the trimer of S2 and fuses the viral and host lipid bilayers which release the viral ribonucleoprotein complex into the cell.[26]
As a result, even if the virus penetrates epithelial cells, the immune system cannot detect it early and suppress it in the first place. Additionally, the slime affinity of these mutated spikes increased by a factor of 50.[27] In general, viruses do not find cells which they directly infect like bacteria do, but when they are poured onto cells in a large amount, some of them travel through mucus in the respiratory tract and sometimes engage epithelial cell receptors and spikes, which leads to infection.[28] The increased mucus tropism means that infection can easily be initiated by far fewer viruses than the former prototype.[27, 29]
3. The risk factors of COVID-19
Table 1 and Fig. 3 shows the number of infected cases, the number of deaths, and the mortality rate by age group in South Korea since October 27, 2022. The age group over 80 years old had the highest number of deaths as well as the highest mortality rate, meaning the ratio of the number of deaths of the number of infected people.
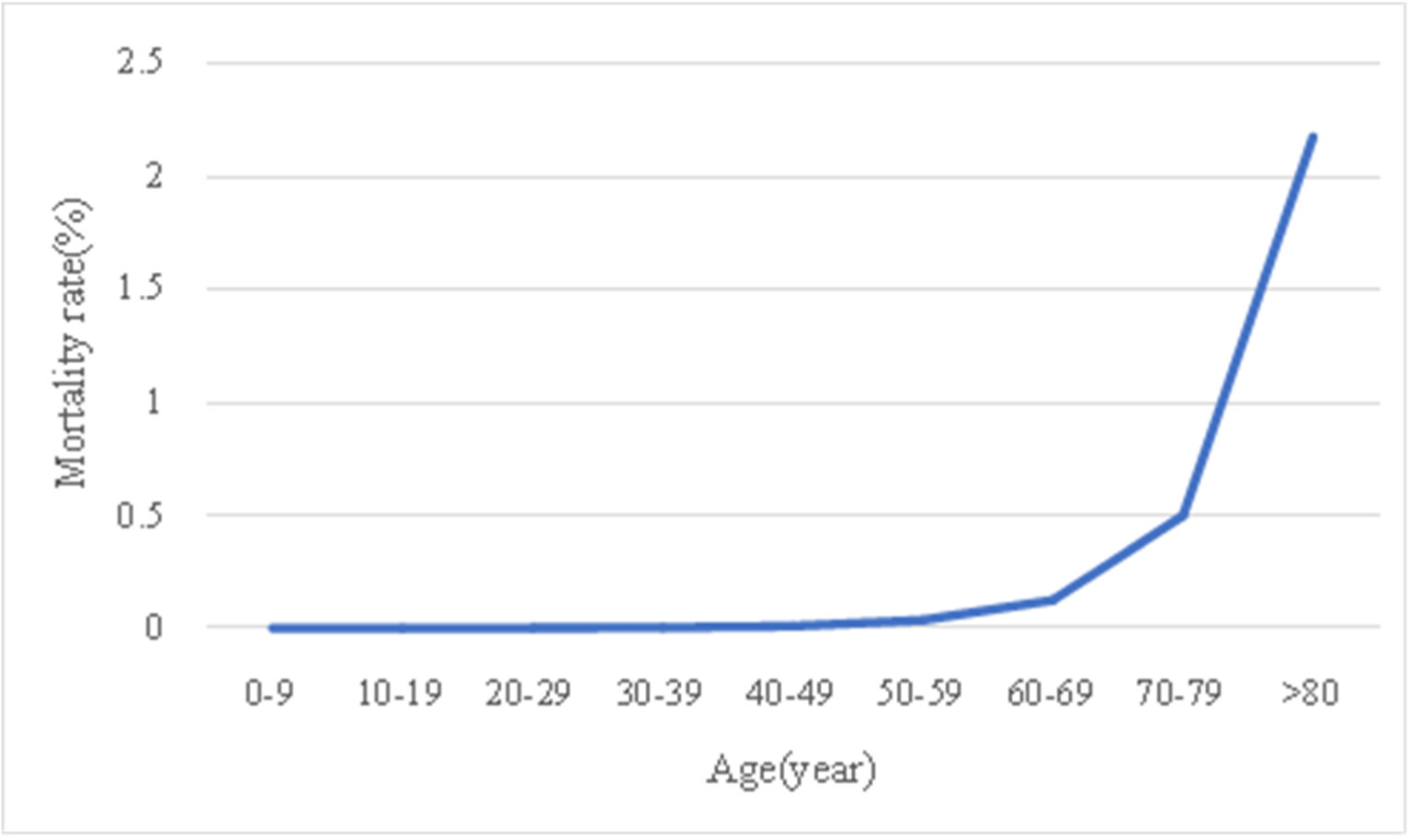
According to Figure 3, a graph showing the mortality rate for each age group, it can be found that the mortality rate generally increases with the age. In particular, it shows that the mortality rate increases sharply in the age group after 50-59. Also, comparing the ratio of the number of deaths in each age group to the total number of deaths, the over 80 age group account for 59.1% and the70-70 age group account for 23.0%. This indicates that 82.1% of the deaths are over 70 years old.
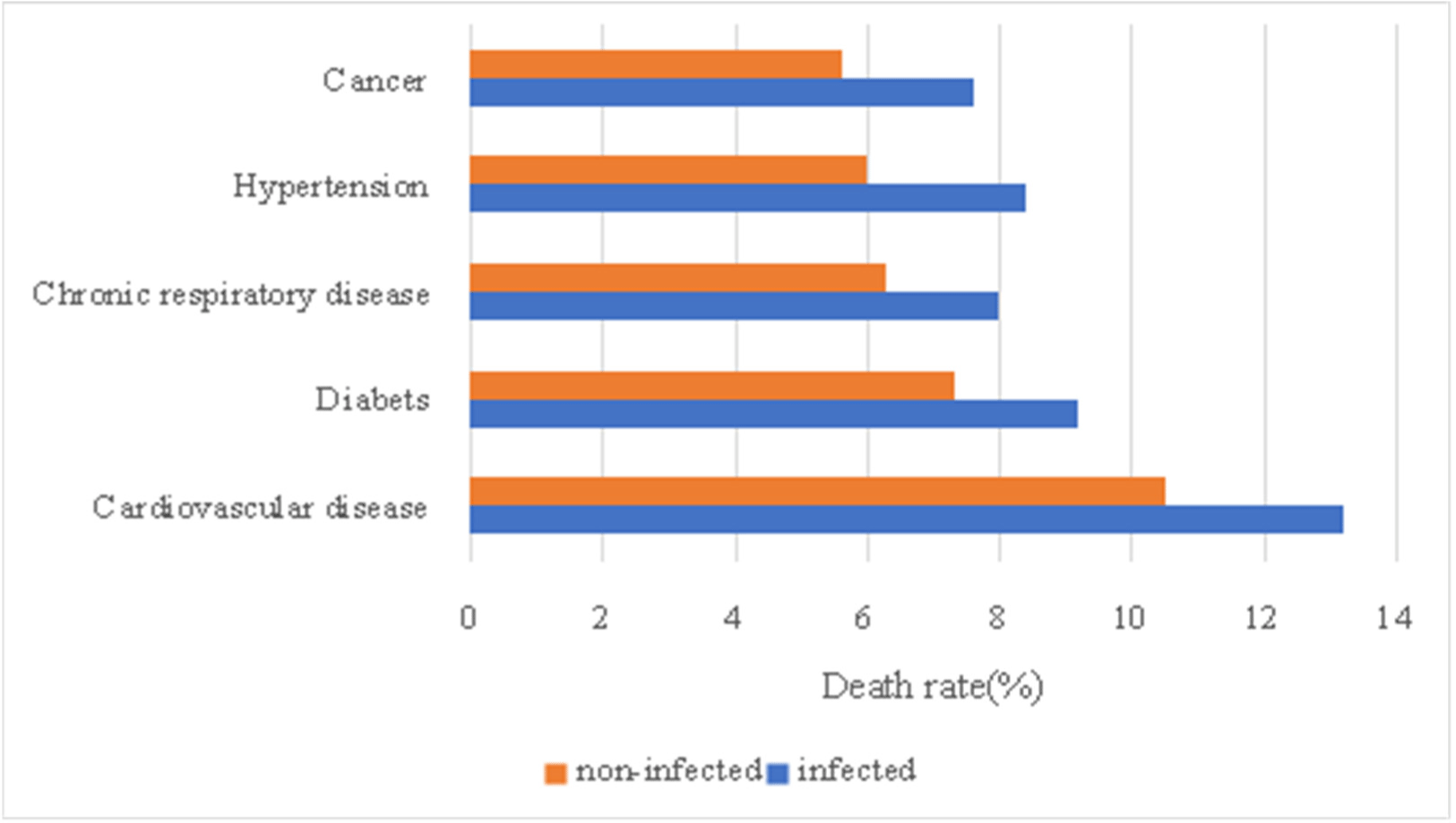
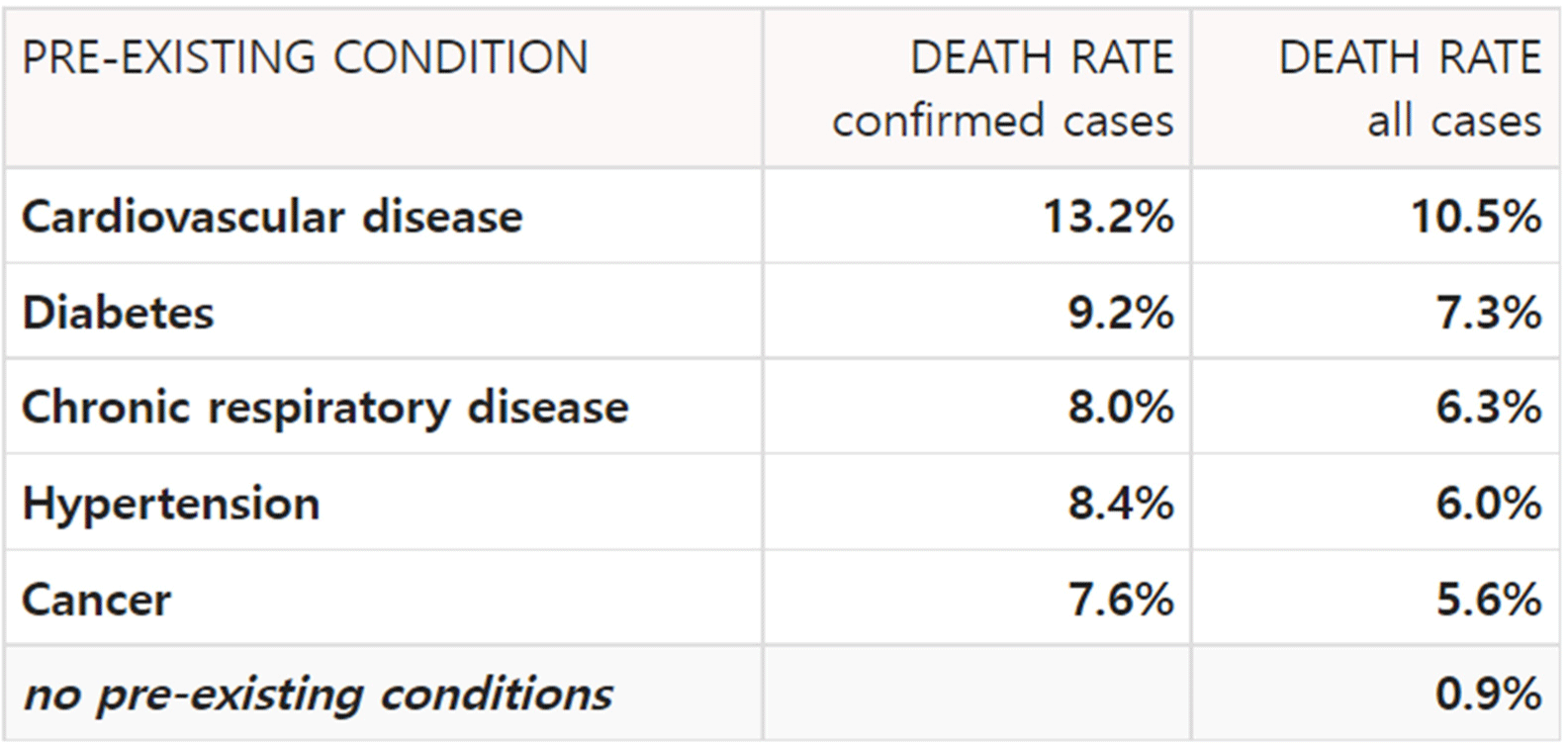
Fig. 4 and Fig. 5 is table of cases in which mortality increases due to diseases before becoming infected with SARS-CoV-2. First, for cardiovascular disease, mortality rate increased from 10.5% to 13.2%. In the case of diabetes, mortality rate increased from 7.3% to 9.2%, and in the case of chronic respiratory disease, mortality rate increased from 6.3% to 8.0%. For hypertension, the mortality rate increased from 6.0% to 8.4%, and in cancer, the mortality rate increased from 5.6% to 7.6%. The rate at which the death rate increased due to SARS-CoV-2 is 25.7%, 26.0%, 26.9%, 40.0%, and 35.7%, so hypertension and cancer can be seen to be the diseases with the greatest increase in mortality due to SARS-CoV-2 infection. In other words, it can be said that the diseases are the biggest risk factor of SARS-CoV-2. The graph of Figure 4 and 5 below summarizes these differences.
4. Conclusion
The occurrence, duration, and severity of these symptoms vary from person to person, which makes symptoms of the COVID-19 infection ranging from those of a mild cold to death in severe cases. The cause of these differences can be found in individual characteristics of the patient. Age, former diseases of the patient, and they medications they took can such characteristics. As the SARS-CoV-2 affects the lung, diseases related to the lung such as chronic obstructive pulmonary disease, lung cancer, cystic fibrosis, pulmonary fibrosis, moderate to severe asthma, pulmonary hypertension, and pulmonary embolism are especially more dangerous to human body when infected by the SARS-CoV-2. Though various methods including comparing the characteristics of mutant viruses and subunits of the COVID-19, we aimed to describe the risk factors and deterioration of COVID-19 and the strain of SARS-CoV-2 during COVID-19 pandemic.