1. Introduction
In November, 2021, the Omicron variant was confirmed among multiple countries and expanded quickly across the globe as a new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).[1] Much consideration has been drawn to scientists that the Omicron variant may become the dominant coronavirus disease over the 2019 (COVID-19) variant at a surprisingly fast rate. This variant is a new considerably mutated SARS-CoV-2 variant (i.e., B.1.1.529) from its original virus structure and was labeled as a variant of concern by the World Health Organization on November 26, 2021, which subsequently grew into the dominant COVID-19 variant across the world.[2]
This review aimed to highlight how the SARS-CoV-2 Omicron variant is genetically different than the other variants, how fast it can disseminate from individual to individual, how efficient the current COVID-19 vaccines are against it, and how we should approach developing a novel vaccine in order to prevent individuals from being infected with it.[3]
2. How is the SARS-CoV-2 Omicron variant genetically different than the other variants?
The Omicron variant of the SARS-CoV-2 (B.1.1.529) genome comprises 18,261 mutations where approximately 97% mutations are existent in the coding region, and the rest of the mutations (i.e., 558) are observed in the extragenic part (Fig. 1).[4] Mutations in the coding region are 2,965 insertions and deletions, and incompatible and compatible single-nucleotide polymorphisms mutations are 11,995 and 2,743, respectively.[4] Thirty mutations have been mainly detected at the receptor-binding domain of the spike (S) protein of the Omicron variant.[5] For over 423 million-detailed SARS-CoV-2 infections across the globe as of Feb 14, 2022, the virus has expanded into more than 1,500 specific phylogenetic assignments of named global outbreak lineages.[6] Further, three other deletions and one insertion mutation is located in the S protein. The Global Initiative on Sharing All Influenza 101 Data demonstrated preliminary data indicating that the N-Terminal Domain consists of 11 mutations, comprising six deletions and one insertion, including mutations N211 and ins214EPE exclusively observed in the Omicron variant.[7]
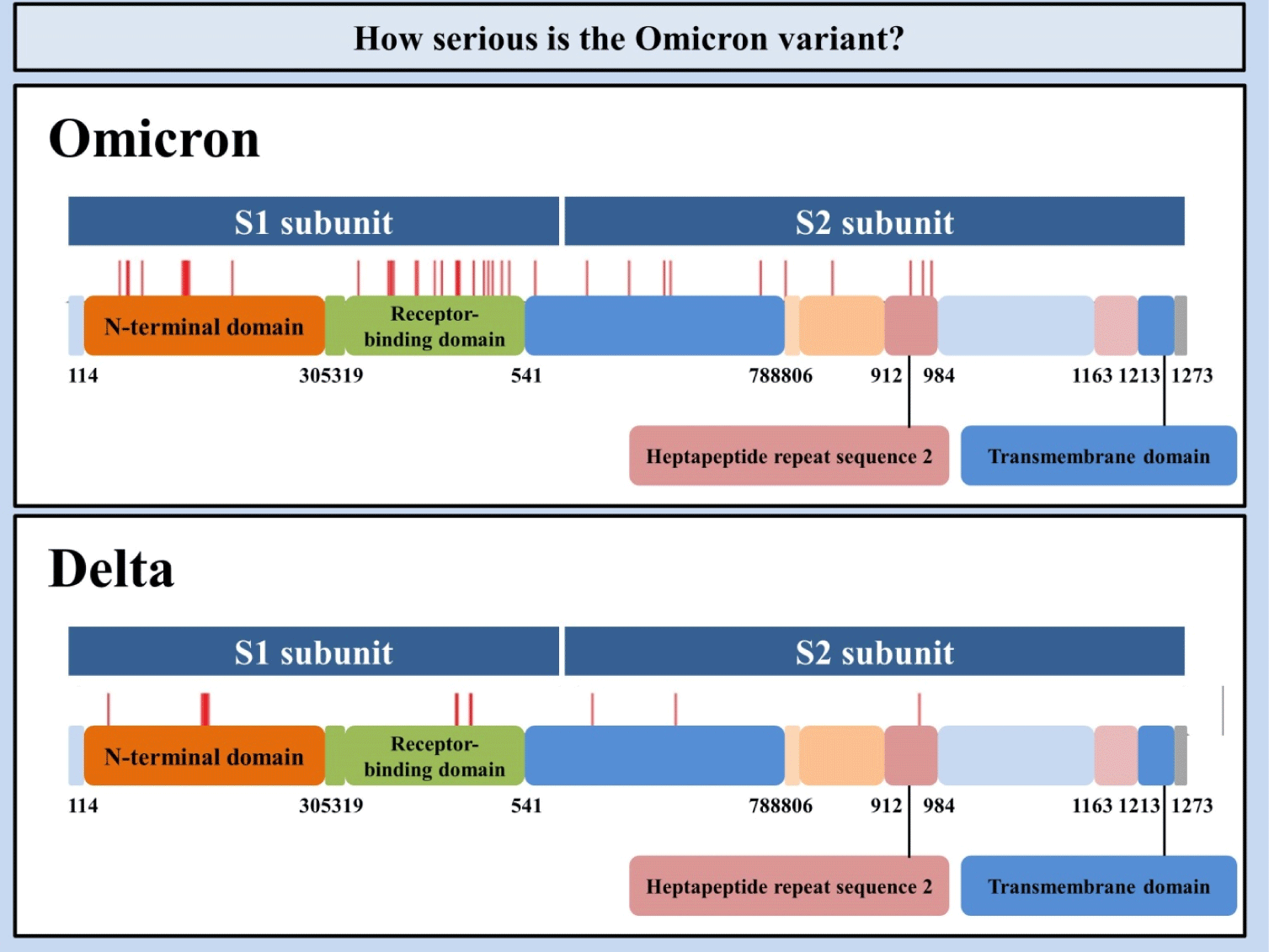
Much to our surprise, some mutations were previously detected in the preceding virus of concern that helped neutralizing antibodies.[8] Five distinct variants of SARS-CoV-2 have been recognized as viruses of concern at various time points.[4] It has been suggested that 4 mutations (i.e., N501Y, D614G, K417N, and T478K) together with novel mutations existent in the Omicron variant have less resistance to vaccines already in the market and boost the risk of re-infectivity.[8, 9] Regarding the Delta and Omicron variants’ mutations, the mutations mentioned above have two out of three receptor-binding domain mutations in common.[9] The first mutation, a lysine to asparagine change at position 417, has been associated with the S protein architectural amendments that may augment immune evasion. The second mutation, a threonine to lysine change at position 478, is prone to promoting the residue's electrostatic potential and steric interference, as it may be increasing receptor-binding domain binding affinity and granting immunological escape. A leucine to arginine change at position 452 is detected in Delta, but not in the Omicron variant. It is perceived to increment affinity for angiotensin-converting enzyme 2 (ACE2) receptors identified on the surface of diverse human cells, encompassing the lungs.[10] It is recognized that Wuhan-Hu-1 has 1273 amino acids, the Delta variation 1271, and the Omicron variant 1270.[11] The reason that the Delta and the Omicron variants have lesser residues than the wild-type is ascribed to their sequence loss. Genome analyses showed the Omicron variant of SARS-CoV-2 structures as a novel monogenetic group.[12] Besides, scientists reported that the Omicron variant of SARS-CoV-2 was derived from the 20B clade and constituted two subclades.[13]
3. How fast is the Omicron variant spreading in the world?
Concerns of the Omicron variant infectivity were raised as it spread across the world and the number of cases have increased dramatically.[14] Notably, it has become the dominant variant worldwide within 3 months of its emergence. A study from Britain reported that the number of subjects who were infected with the Omicron variant doubled every 2-3 days.[15] Scientists applied the linear regression of each pseudovirus to compare them with the wild type across the full scope and concluded that the Delta variant was almost two-fold more effective at inflicting target cells, the Gamma variant had similar transmissibility, and the Beta variant had less infectivity when compared to the wild type.[6] Of great clinical importance, the Omicron variant had an infection rate four times greater than the wild type and two times greater than the Delta variant.[6] These observations demonstrate that spike sequence plays a critical part in transmissibility, with the Omicron variant exhibiting more efficient ACE2-mediated infection than the wild type or other variants.[6]
Various factors may affect the escalated transmissibility rate of the Omicron variant. The most important factor is approximately 30 mutations in the spike protein that the SARS-CoV-2 protein utilizes when detecting host cells.[16] A previous study on these mutations describes the likelihood of the high infectivity rate by escaping the immune reactions.[8, 17] For instance, the N501Y mutation of the Omicron variant boosts the binding affinity with the ACE2 receptor which plays an important part in the transmission process. Further, the binding affinity becomes more secure in conjunction with Q498R, which results in better accessibility of the Omicron variant into the host.[8, 13] Moreover, evidence reporting that patients formerly infected with COVID-19 are vulnerable to re-infection of the Omicron variant indicates that the Omicron has high immune evasion capability and higher infectivity.[8] Mutations recognized in the Omicron variant (i.e., H655Y and N679K) are detected adjacent to the furin cleavage site and may increase spike cleavage, leading to the virus more infective at a population level,[18] while P681H can magnify infectivity by augmenting the spike protein cleavage.
Additionally, the Omicron variant may yield a false negative result in polymerase chain reaction tests because of the “S gene target failure,” which then gives the way of transmitting the virus infection at an astonishingly high rate across the world without being identified.[19] A previous research work reported a plausible relationship between the positive electrostatic potential and affinity in the Delta virus of concern.[20] The escalated electrostatic potential is acknowledged in the case of Delta and Delta-plus variants of SARS-CoV-2, incorporating the Omicron variant at the receptor-binding domain interface with ACE2.[21] The titers of various pseudotyped SARS-CoV-2 S/HIV-1 viruses were assessed using HEK293T cells stably expressing the ACE2 receptor. Omicron variant S/HIV-1 pseudotyped viruses definitely require entering the HEK293T with the help of AGE2. The receptor-binding domain and ACE2 preserve a nanomolar level of binding affinity, which is comparable among the Beta, the Delta, and the Omicron. Because ACE2 is needed for the receptor-binding domain, it seems that all variants have previously extended to the nanomolar scale, causing problems for the virus to advance further.[21] Another piece of research work concluded that the Omicron variant had boosted affinity to the ACE2 when compared with the other SARS-CoV-2 variant.[22] Many mutations in the receptor-binding domain of spike protein of Omicron variants, including Q493R, N501Y, S371L, S373P, S375F, Q498R, and T478K, are culpable for the higher affinity to the ACE2,[11, 13] which all indicate that the Omicron variant has higher infectivity than the other variants.[21]
4. How efficient are the current COVID-19 vaccines in opposition to the SARS-CoV-2 Omicron variant?
Several scientists evaluated the mutations detected in the receptor-binding domain of the spike of the Omicron variant and came to the result that the utilization of the coronavirus-specific attachment inhibitors is less efficient compared to the past. Based on the fact that the Omicron variant of SARS-CoV-2 was detected in COVID-19 vaccinated subjects, it can be reassured that it is crucial to develop vaccines that will work efficiently to protect individuals from the variant.[3] With regard to such observations and the following new demand, previous studies have proven that the mRNA vaccine had decreased neutralization effects against the Omicron variant; however, they still had significant neutralization for this novel variant.[23] This may, in part, be ascribed to the fact that mRNA vaccines, including Ad26.COV2.S and BNT162b2 vaccines, had lasting spike-specific CD8+ and CD4+ T cell reactions, a finding that demonstrated comprehensive cross-reactivity against both of the Delta and the Omicron variants, comprising in central and effector memory cellular sub-communities. Median levels of the Omicron spike-specific CD8+ T cell reactions were estimated at 82–84% of the original SARS-CoV-2 WA1/2020 spike-specific CD8+ T cell reactions. This finding provides immunological evidence for the hypothesis that the vaccines currently in use still demonstrate sturdy protection against serious disease with the SARS-CoV-2 Omicron variant in spite of the significantly diminished neutralizing antibody reactions.[24] In addition, a recently published paper demonstrated that primary vaccination with two doses of AstraZeneca or Pfizer/BioNTech vaccine yielded lower safety against symptomatic disease induced by the Omicron variant.[25] They also showed that Pfizer or mRNA-1273 booster shots after either AstraZeneca or Pfizer primary course significantly escalated protection; however, that protection declined over time.[26] In corresponding well with this finding, recent studies determined that an mRNA booster is critical in neutralizing the Omicron variant.[6, 27] In coping with the Omicron variant in clinical settings, Pfizer-BioNTech has stated that they could produce vaccines specific to the Omicron by April 2022.[26] Moderna has also been developing an updated vaccine efficient against the Omicron variant and stated that it could be completed with examination and available with the regulators on a similar timescale.[28] However, neither of them has announced any significant news regarding this matter as of Feb 20, 2022. A vaccine derived from the Omicron would seemingly require only two doses to securely obtain efficiency for the variant.[2] Until the novel vaccines for the Omicron are available on the market, a booster shot may be the only option. Even after the novel vaccines for the Omicron are developed, it would be a great economical and societal burden on public health of undeveloped countries in the sense that they would still have to provide their citizens with new vaccines for the Omicron variant while a great portion of them cannot have access to out-of-date vaccines and that the virus is prone to mutations in countries with low vaccination rates and high transmissibility rates.[29]
5. How can the ‘Stealth’ Omicron, Omicron sublineage BA.2, evade vaccination and how are they different from the Omicron variant itself?
The SARS-CoV-2 B.1.1.529 (Omicron) variant has 3 main sublineages: BA.1, BA.2, and BA.3.[2] When compared with the Wuhan/Hu-1/2019 reference strain, the sublineage BA.2 of the Omicron variant comprises 16 amino acid substitutions in the receptor-binding domain of the S protein of SARS-CoV-2, which is the main focal point for monoclonal antibody-based treatment.[2] The BA.2 and BA.1 variants have 12 of these 16 substitutions in common; however, BA.2 has 4 changes in the receptor-binding domain (i.e., S371F, T376A, D405N, and R408S) which were found to be different from those in BA.1, a finding indicating that there could be discrepancies in the efficiency of monoclonal antibodies against these distinct Omicron subvariants.[2] One critical difference between BA.2 and BA.1 is that sublineage BA.2 has a defect in the spike gene deletion in the part ciphering amino acid 69/70, which indicates that it will not be determined by the S Gene Target Failure assay, a test utilized as a proxy marker for the Omicron virus of concern.[2] Therefore, it is often indicated as the ‘Stealth’ Omicron variant.[2]







