1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a representative of the Coronaviridae family, which constitutes various virulent strains that transmit to humans and animals, comprising SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV).[1] As of 4 March 2022, the coronavirus disease 2019 (COVID-19) pandemic, caused by SARS-CoV-2, has expanded to 220 countries or areas with approximately 440.8 million laboratory-verified cases and 5.98 million deaths (World Health Organization [WHO]);[2] thus, resulting in global social disruption and economic decline. Progress towards availability of secure and efficient vaccines affordable in all countries and areas is essential to put a halt to this pandemic.
A vaccine is essential, particularly, due to the fact that studies have suggested asymptomatic transmission of COVID-19.[3] The expeditious development, circulation, and administration of a vaccine to the population across the world are the most efficient ways to extinguish the pandemic.[4] In general, an effective vaccine takes decades to develop; therefore, rapid availability of effective vaccines for widespread distribution is extraordinary and may yield unpredicted side effects. Much to the public surprise, novel mass-producing platforms, formation-supported virus antigen composition, computational biology, protein manufacturing, and gene integration all have contributed to the advancement of the tools required to produce vaccines with expedition and accuracy.[5]
In order to establish vaccines against SARS-CoV-2, it is of great scientific importance to appropriately interpret the immune systems involved in protection of subjects against the virus. Mounting evidence has shown the value of both humoral and cellular immunity with regard to the development of a protective effect against COVID-19.[6] For instance, neutralizing antibodies (nAbs) have been reported to reduce the amount of the virus, with a greater response among subjects whose immune system had not yet been infected or who had a high amount of the virus from the beginning.[6] In conjunction with nAbs, T cell responses have an essential role in COVID-19 disease. For example, a study of patients affected by COVID-19 pneumonia showed markedly increased capability of CD4+ and CD8+ T cells to produce in vitro interleukin-17, thereby enhancing the inflammatory response and stimulating neutrophils.[7] Thus, it can be inferred that ideal vaccines should induce both the humoral and cellular immune-mediated reactions in a similar way to the virus that infects the subject in a real world setting.
With respect to global demand for effective vaccines against COVID-19, researchers across the world are in the race of developing COVID-19 vaccines. In accordance with the COVID-19 vaccine tracker and landscape by the WHO, COVID-19 vaccine contenders are classified into 7 categories (Fig. 1), which may be divided into 3 classes: first, protein-based vaccines that use major antigens (i.e., inactivated virus vaccines, virus-like particles [VLPs], and protein subunit vaccines); second, gene-based vaccines that transport genes encoding viral antigens in host cells for in vivo manufacturing (i.e., virus-vectored vaccines, DNA vaccines, and mRNA vaccines); and, last, a compound of both protein-based and gene-based access to yield protein immunogens both in vitro and in vivo (i.e., live-attenuated viral vaccines). As of 4 March, 2022, the WHO detailed 147 vaccines in clinical development including 32 in Phase 3 trials and 10 in Phase 4 trials (WHO; COVID-19 Vaccine tracker). In this review article, we outline and review the targeting spots of utmost scientific value in vaccine contenders with a special focus on the vaccines in use with reported data.
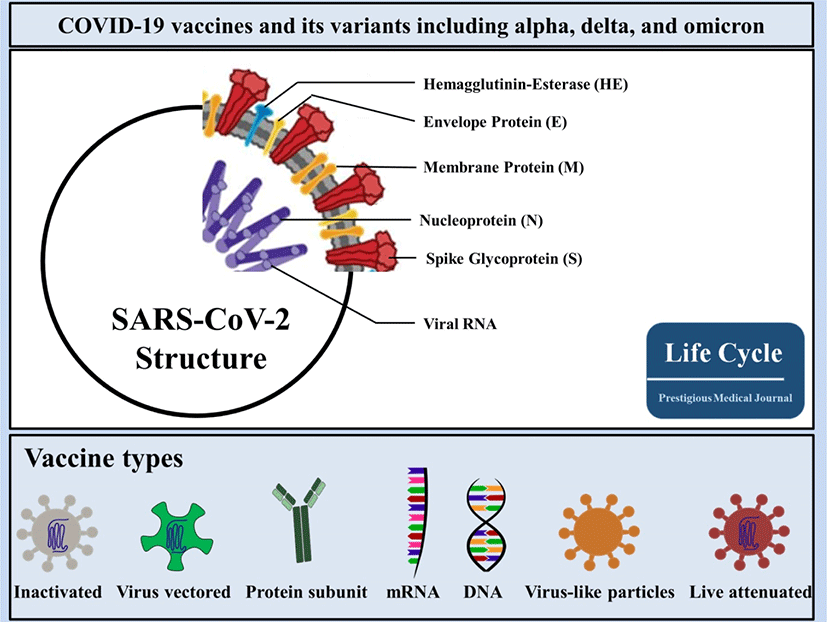
2. Various SARS-CoV-2 molecular proteins as targeting spots for vaccines
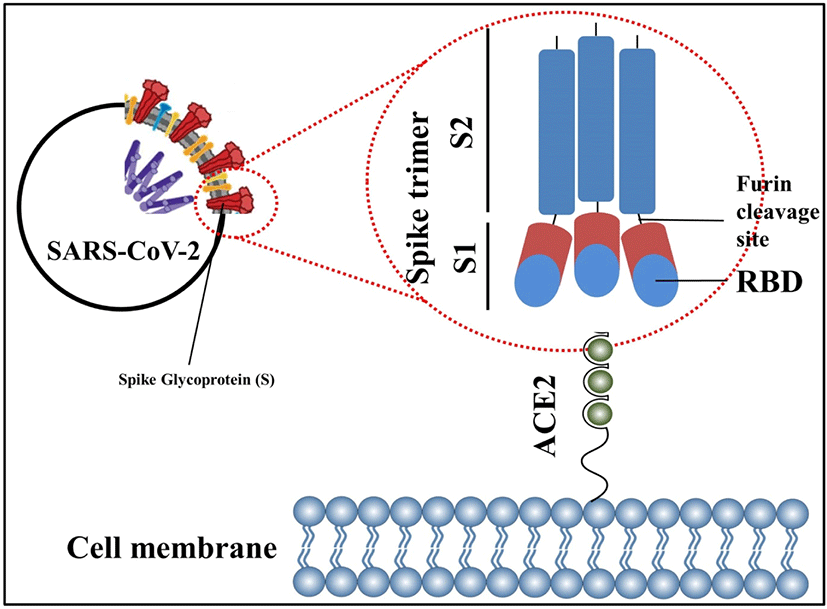
SARS-CoV-2 comprises 4 main structural proteins, specifically, 1) spike (S), 2) membrane (M) and 3) envelope (E) proteins; all of them are enclosed in the viral outer surface envelope, and 4) nucleocapsid (N) protein, which is located in the ribonucleoprotein core (Fig. 1).[8] S protein is considered the major protein utilized as a target point in the progress of COVID-19 vaccine development. It is divided into two critical functional subunits; the first one is a membrane-distal S1 subunit composing the globular head of the S protein; and the other one is a membrane-proximal S2 subunit comprising the stalk of the protein. Once engaged with a host cell, the S1 subunit of the virus recognizes and attaches to receptors on the host cell through its receptor-binding domain (RBD); subsequently, the S2 subunit on the virus fuses the envelope of the virus with the host cell membrane, which is a vital process for viral entrance (Fig. 2). In MERS-CoV, SARS-CoV, or SARS-CoV-2, the RBD is positioned in the C-terminal domain of the S1 subunit.[9] In a few coronaviruses, the N-terminal domain (NTD) of the S1 subunit may become utilized for receptor binding. The S2 subunit consists of the fusion peptide (FP), connecting region (CR), heptad repeat 1 (HR1) and HR2 in the vicinity of a central helix as a helix-turn-helix formation. Structural information has reported an image for the realignment of SARS-CoV-2 S molecular protein when engaging with the host cell receptor.[10]. The binding of the RBD to its cellular receptor, human angiotensin-converting enzyme 2 (hACE2), induces the disconnection of the S1 subunit and simultaneously institutes the re-bending transformation of the spring-bundled S2 subunit, that bulges the FP at its end for the membrane fusion process. The S2 subunit in its post-fusion configuration convolutes in the formation of a lengthy helical array adjacent to the FP embedded into the host cell membrane. Hypothetically, nAbs can aim at the S protein in order to hinder viral infection at various phases in the course of viral entry. The RBD is the main spot where nAbs hamper viral receptor binding.[11] Further, the S2 subunit is a possible point for nAbs that impede structural adjustment of the S protein and the embedment of FP essential for virus-host membrane fusion.[12] Furthermore, the S protein is a major spot for eliciting T cell reactions, which is supported by previous studies detailing both CD4+ and CD8+ T cell epitopes in the S protein of SARS-CoV-2.[13]
3. Virus-vectored and mRNA vaccines for the prevention of SARS-CoV-2
Up to now, three COVID-19 vaccines established upon adenovirus vector with the full-length S protein on the surface have been approved for use; the first one is based on a single-dose of human adenovirus type 5 (Ad5) (i.e., CanSino Biological Inc./Beijing Institute of Biotechnology),[14, 15] the second one uses two-doses of recombinant chimpanzee adenovirus, ChAdOx1-S,(i.e., AstraZeneca/University of Oxford)[16, 17] and the third one is a single-dose Ad26.COV2.S vaccine.[18] There are three other vaccines using adenovirus in Phase 3 trials: Recombinant COVID-19 vaccine (adenovirus type 5 vector) for inhalation (Ad5-nCoV-IH),[19] combination of human Ad26 and Ad5 in a prime-boost vaccination regimen (Gam-COVID-Vac Adeno-based [rAd26-S+rAd5-S], Sputnik V),[20] and DeINS1-2019-nCoV-RBD-OPT1 (Intranasal flu-based-RBD). While adenoviral vectors have the advantages of their broad tissue tropism, intrinsic adjuvant elements, and scalability, they also have disadvantages of pre-existing immunity in humans, which may decrease efficiency of the adenoviral vector.[4] To date, there are two DNA vaccine contenders in Phase 3 trial[21] and none in Phase 4 trials.
The S protein is labile when used as a recombinant molecular protein and is likely to reconfigure from its pre-fusion to a post-fusion arrangement, discharging the S1 subunit (Fig. 2). Nonetheless, the S1 subunit is the immunologically predominant immunogen when infected with CoV because of its convenience for immunological perception and its consisting of neutralizing epitopes primarily on its RBD.[13] Methods to preserve the S protein in its pre-fusion arrangement and to augment pre-fusion S protein expression are considered to boost the aspect and capacity of vaccine-induced antibodies pointing at the practically compatible epitopes on the surface of the S1 subunit. Two proline substitutions (2P) at the pinnacle of the central helix and HR1 may hold the S proteins of MERS-CoV, SARS-CoV, and coronavirus HKU1 in the immunologically ideal pre-fusion arrangements (Fig. 2). The ensuing antigen, S-2P, generated higher nAb titers than wild-type S protein in animal models.[22] Based on the earlier encounters with the CoVs, the S-2P arrangement is currently utilized in some vaccine approaches against COVID-19. SARS-CoV-2 S-2P (constituting proline substitutions at residues K986 and V987) is employed as the major immunogen in three gene-based vaccines (i.e., mRNA vaccines by Moderna/National Institute of Allergy and Infectious Diseases [NIAID] administered two-doses with 28 days apart[23] and BioNTech/Pfizer given two-doses 21 days apart[24], and a recombinant Ad26 vaccine manufactured by Janssen Pharmaceutical Companies delivered as a singular dose[18]) (Table 1, Fig. 1). Further, mutations at the separation spots in the S protein are considered to preserve the pre-fusion arrangement of the S protein. S-2P in the Janssen Ad26-vectored vaccine (Ad26.COV2.S) consists of increased mutations at the S1–S2 polybasic breaking spot from RRAR to SRAG or QQAQ to deliver it protease resistant, which aids to further sustain the S protein in its pre-fusion configuration.[25] All of the aforementioned vaccines, which are in Phase 3 and 4, evoked abundant protective immunity against COVID-19 (Table 1). Of the vaccines mentioned above, the research papers for the two mRNA vaccines (BNT162b2 by BioNTech/Pfizer and mRNA1273 by Moderna/NIAID) and the protein subunit vaccine, NVX-CoV2373[26], inspiringly detailed that these vaccines evoked both great titers of nAbs in recuperating subjects with COVID-19 infection and extensive T cell reactions. Hence, maintaining the S protein in its pre-fusion arrangement displays an efficient route to enhance vaccine competence. Further, DNA or RNA vaccines have distinct advantages that as well as antibody and CD4+ T cell responses, they evoke CD8+ cytotoxic T cell reactions, which play a critical part for virus eradication.[21] It is of great clinical significance to note that clinical studies by BioNTech/Pfizer and Moderna/NIAID have reported that their mRNA vaccines have approximately 95% effectiveness in the prevention of COVID-19.[23]
| ID | Vaccine platform description | Type of contender vaccine | Number of doses | Schedule | Route of administration | Developers | Phase | References |
|---|---|---|---|---|---|---|---|---|
| 1 | Inactivated virus | CoronaVac; inactivated SARS-CoV-2 vaccine (vero cell) | 2 | Day 0 + 14 | IM | Sinovac Research and Development Co., Ltd | Phase 4 | [32] |
| 2 | Inactivated virus | Inactivated SARS-CoV-2 vaccine (Vero cell) | 2 | Day 0 + 21 | IM | Sinopharm; | Phase 4 | A |
| 3 | Inactivated virus | Inactivated SARS-CoV-2 vaccine (Vero cell), vaccine name BBIBP-CorV | 2 | Day 0 + 21 | IM | Sinopharm; | Phase 4 | [31] |
| 14 | Inactivated virus | SARS-CoV-2 vaccine (vero cells) | 2 | Day 0 + 28 | IM | Institute of Medical Biology + Chinese Academy of Medical Sciences | Phase 3 | B |
| 15 | Inactivated virus | QazCovid-in® - COVID-19 inactivated vaccine | 2 | Day 0 + 21 | IM | Research Institute for Biological Safety Problems, Rep of Kazakhstan | Phase 3 | C |
| 20 | Inactivated virus | BBV152 vaccine | 2 | Day 0 + 14 | IM | Bharat Biotech International Limited | Phase 3 | D |
| 25 | Inactivated virus | Inactivated SARS-CoV-2 vaccine (Vero cell) | 2 | Day 0 + 28 | IM | Shenzhen Kangtai Biological Products Co., Ltd. | Phase 3 | E |
| 56 | Inactivated virus | VLA2001 | 2 | Day 0 + 21 | IM | Valneva, National Institute for Health Research, United Kingdom | Phase 3 | F |
| 63 | Inactivated virus | TURKOVAC, inactivated virus | 2 | Day 0 + 21 | IM | Erciyes University and the Health Institutes of Turkey (TUSEB) | Phase 3 | G |
| 4 | Viral vector (non-replicating) | ChAdOx1-S - (AZD1222) | 1-2 | Day 0 + 28 | IM | AstraZeneca + University of Oxford | Phase 4 | [15] |
| 5 | Viral vector (non -replicating) | Recombinant novel coronavirus vaccine (adenovirus type 5 vector) | 1 | Day 0 | IM | CanSino Biological Inc./Beijing Institute of Biotechnology | Phase 4 | [13, 14] |
| 6 | Viral vector (non -replicating) | Recombinant COVID-19 vaccine (adenovirus type 5 vector) for Inhalation (Ad5-nCoV-IH) | 1 | Day 0 | IH | CanSino Biological Inc./Beijing Institute of Biotechnology | Phase 3 | H |
| 7 | Viral vector (non -replicating) | Gam-COVID-Vac Adeno-based (rAd26-S+rAd5-S) | 2 | Day 0 + 21 | IM | Gamaleya Research Institute ; Health Ministry of the Russian Federation | Phase 3 | [10] |
| 8 | Viral vector (non -replicating) | Ad26.COV2.S | 1-2 | Day 0 or Day 0 +56 | IM | Janssen Pharmaceutical | Phase 4 | [17] |
| 38 | Viral vector (replicating) | DeINS1-2019-nCoV-RBD-OPT1 (Intranasal flu-based-RBD) | 2 | Day 0 + 28 | IN | University of Hong Kong, Xiamen University and Beijing Wantai Biological Pharmacy | Phase 3 | I |
| 9 | Protein subunit | SARS-CoV-2 rS/Matrix M1-Adjuvant (Full length recombinant SARS CoV-2) | 2 | Day 0 + 21 | IM | Novavax | Phase 3 | [25] |
| 12 | Protein subunit | Recombinant SARS-CoV-2 vaccine (CHO cell) | 2-3 | Day 0 + 28 or | IM | Anhui Zhifei Longcom Biopharmaceutical + Institute of Microbiology, Chinese Academy of Sciences | Phase 3 | J |
| 22 | Protein subunit | VAT00008: SARS-CoV-2 S protein with adjuvant | 2 | Day 0 + 21 | IM | Sanofi Pasteur + GSK | Phase 3 | K |
| 29 | Protein subunit | CpG 1018/Alum-adjuvanted recombinant SARS-CoV-2 trimeric S-protein subunit vaccine (SCB-2019) | 2 | Day 0 + 21 | IM | Clover Biopharmaceuticals Inc./Dynavax | Phase 3 | L |
| 30 | Protein subunit | COVAX-19® Recombinant spike protein + adjuvant | 2 | Day 0 + 21 | IM | Vaxine Pty Ltd./CinnaGen Co. | Phase 3 | M |
| 31 | Protein subunit | MVC-COV1901 (Spike-2P protein + adjuvant CpG 1018) | 2 | Day 0 + 28 | IM | Medigen Vaccine Biologics + Dynavax + National Institute of Allergy and Infectious Diseases (NIAID) | Phase 4 | N |
| 33 | Protein subunit | FINLAY-FR-2 anti-SARS-CoV-2 vaccine (RBD chemically conjugated to tetanus toxoid plus adjuvant) | 2 | Day 0 + 28 | IM | Instituto Finlay de Vacunas | Phase 3 | O |
| 34 | Protein subunit | EpiVacCorona (EpiVacCorona vaccine based on peptide antigens for the prevention of COVID-19) | 2 | Day 0 + 21 | IM | Federal Budgetary Research Institution State Research Center of Virology and Biotechnology "Vector" | Phase 3 | P |
| 35 | Protein subunit | RBD (baculovirus production expressed in Sf9 cells) recombinant SARS-CoV-2 vaccine (Sf9 Cell) | 2 | Day 0 + Day 21 + Day 42 | IM | West China Hospital + Sichuan University | Phase 3 | Q |
| 55 | Protein subunit | CIGB-66 (RBD+aluminium hydroxide) | 3 | Day 0 + 14 + 28 | IM | Center for Genetic Engineering and Biotechnology (CIGB) | Phase 3 | R |
| 57 | Protein subunit | BECOV2 | 2 | Day 0 + 28 | IM | Biological E. Limited | Phase 3 | S |
| 60 | Protein subunit | Recombinant Sars-CoV-2 spike protein, aluminum adjuvanted (Nanocovax) | 2 | Day 0 + 21 | IM | Nanogen Pharmaceutical Biotechnology | Phase 3 | T |
| 61 | Protein subunit | Recombinant protein vaccine S-268019 (using Baculovirus expression vector system) | 2 | Day 0 + 21 | IM | Shionogi | Phase 3 | U |
| 65 | Protein subunit | GBP510, a recombinant surface protein vaccine with adjuvant AS03 (aluminium hydroxide) | 2 | Day 0 + 28 | IM | SK Bioscience Co., Ltd. and CEPI | Phase 3 | V |
| 66 | Protein subunit | Razi Cov Pars, recombinant spike protein | 3 | Day 0 + 21 +51 | IM and IN | Razi Vaccine and Serum Research Institute | Phase 3 | W |
| 91 | Protein subunit | Recombinant SARS-CoV-2 fusion protein vaccine (V-01) | 2 | Day 0 + 21 | IM | Livzon Pharmaceutical | Phase 3 | X |
| 106 | Protein subunit | RBD protein recombinant SARS-CoV-2 vaccine (Noora vaccine) | 3 | Day 0 + 21 + 35 | IM | Bagheiat-allah University of Medical Sciences/AmitisGen | Phase 3 | Y |
| 10 | RNA based vaccine | mRNA-1273 | 2 | Day 0 + 28 | IM | Moderna + National Institute of Allergy and Infectious Diseases (NIAID) | Phase 4 | [22] |
| 11 | RNA based vaccine | BNT162b2 (3 LNP-mRNAs ), also known as "Comirnaty" | 2 | Day 0 + 21 | IM | Pfizer/BioNTech + Fosun Pharma | Phase 4 | [23] |
| 13 | RNA based vaccine | CVnCoV vaccine | 2 | Day 0 + 28 | IM | CureVac AG | Phase 3 | Z |
| 40 | RNA based vaccine | SARS-CoV-2 mRNA vaccine (ARCoV) | 2 | Day 0 + 14 or Day 0 + 28 |
IM | Academy of Military Science (AMS), Walvax Biotechnology and Suzhou Abogen Biosciences | Phase 3 | AA |
| 78 | RNA based vaccine | mRNA-1273.351. | 3 | Day 0 or Day 0 + 28 | IM | Moderna + National Institute of Allergy and Infectious Diseases (NIAID) | Phase 4 | BB |
| 115 | RNA based vaccine | ARCT-154 mRNA vaccine | 2 | Day 0 + 28 | IM | Arcturus Therapeutics, Inc. | Phase 3 | CC |
| 16 | DNA based vaccine | INO-4800+electroporation | 2 | Day 0 + 28 | ID | Inovio Pharmaceuticals + International Vaccine Institute + Advaccine (Suzhou) Biopharmaceutical Co., Ltd | Phase 3 | [20] |
| 18 | DNA based vaccine | nCov vaccine | 3 | Day 0 + 28 + 56 | ID | Zydus Cadila | Phase 3 | DD |
| 41 | Virus like particle | Coronavirus-Like Particle COVID-19 (CoVLP) | 2 | Day 0 + 21 | IM | Medicago Inc. | Phase 3 | EE |
| 53 | Live attenuated virus | COVI-VAC | 1-2 | Day 0 or Day 0 + 28 |
IN | Codagenix/Serum Institute of India | Phase 3 | CC |
APC, antigen presenting cell; ID, intradermal; IM, intramuscular; IN, intranasal; IV, inactivated virus; LAV, live-attenuated virus; PS, protein subunit; VVnr, viral vector (non-replicating); VLP, virus like particle; VVr, viral vector (replicating).
I. Chinese Clinical Trial Register (ChiCTR) - The world health organization international clinical trials registered organization registered platform
M. https://en.irct.ir/trial/57559 2021
The RBD attaches to the host receptor through a receptor-binding motif (RBM) on its external subdomain in SARS-CoV-2.[9] Since glycans cover the surface of the S protein, with the exception of the RBD, and shield it from antibodies, most SARS-CoV-2 nAbs attach to the RBD and prevent the RBD–hACE2 combination, thereby reducing viral adherence.[27] In this regard, there are a handful of clinical trials using RBD-targeted vaccines in Phase 3 (Fig. 1).[28]
In addition, the S1-NTD consists of epitopes for CoV nAbs detected in recovering subjects with COVID-19 and has been recognized as a possible major targeting point in CoV vaccines.[29] NTD-pointing nAbs typically do not explicitly hinder receptor binding; instead, they hamper receptor binding or constrain the S protein structural shifts necessary for the pre-fusion to post-fusion conversion. However, SARS-CoV-2 NTD-binding nAbs typically show less neutralizing potency compared to RBD-specific nAbs.[12] In this regard, NTD-targeting vaccines against COVID-19 have not been developed.
With regard to the S2 subunit, although there is some evidence reporting the S2 subunit as a targeting point for COVID-19 vaccine, S2 subunit-targeting antibodies retrieved from recovering patients reported less neutralizing activities against SARS-CoV-2 than RBD-targeting antibodies.[30]
CoV M and E proteins are hardly able to produce humoral responses, likely as a result of their small molecular sizes and limited ectodomains, which could be recognized by immune cells.[12] (Fig. 2) Although there is some scientific evidence that the sequence identity of M or E proteins in SARS-CoV-2 is considerably higher than for the S protein and RBD, proposing the potential of M and E proteins as targeting points for cross-reactive T cells,[12] there have been no solid studies aiming to develop vaccines using the M or E proteins. The N protein is the most abundant viral protein and is able to produce an immune response following CoV infections.[31] Hence, it is a main target protein not only for evoking antibody reactions, but also for T cell responses.[31] Despite that some scientific evidence shows that N-specific antibodies were found to defend mice against mouse hepatitis virus, a mouse CoV, via Fc-mediated effector functions, anti-N immune responses were not able to show protection against SARS-CoV-2 infection in animal models.[31] Taken together, there is one vaccine using a protein subunit in Phase 4 (i.e., MVC-COV1901 [Spike-2P protein + adjuvant CpG 1018]) and 17 vaccines in Phase 3.[26]
Inactivated virus and live-attenuated virus vaccines utilize the whole virus as a vaccine target. They encompass complete structural proteins (i.e., S, M, E, and N proteins) and vaccines using live-attenuated virus may develop non-architectural and component proteins in vivo. Hence, these vaccine contenders are able to evoke greater humoral and T cell responses than the aforementioned ones, that are established upon a singular protein or protein particles. There are three vaccines using inactivated virus in Phases 4; first, inactivated SARS-CoV-2 vaccine (Vero cell), vaccine named BBIBP-CorV,[32] second, CoronaVac; inactivated SARS-CoV-2 vaccine (Vero cell),[33] and third, inactivated SARS-CoV-2 vaccine (Vero cell) and six others in Phase 3.[34] Since these vaccine formulations are inadequate for replication and safer than live-attenuated vaccines, their inactivation leads to decreased immunogenicity and the need for multiple-dose regimens in order to institute resilient immunity. In addition, they frequently need adjuvants to vaccinate the aging population by reason of immune senescence.[35] In contrast to inactivated virus, live-attenuated vaccines (LAV) are reproducing, but avirulent viruses. LAV administration requires a single-dose to generate immunity. LAVs have the risk of transferring the virus and/or conversion to a pathogenic variety, reactivation in immune-compromised subjects or recombination with alike-viruses disseminating in the general population.[5] Thus far, there has been one SARS-CoV-2 live-attenuated vaccine in Phase 3.[35]
4. Conclusions
To date, several vaccines for SARS-CoV-2 and its variants including alpha, delta, and omicron are available with safety and scientifically reasonable levels of protection efficacy for COVID-19 (i.e., BioNTech/Pfizer and Moderna/NIAID), although more needs to be studied with regard to the effect of mRNA vaccines on the omicron variant. Of note, it is challenging, in a clinical setting, to accurately analyze the effectiveness and the duration of protection by the various vaccines due to the fact that there are no standardized methods to measure neutralization levels. In addition, researchers and the world should find an expeditious way to discover novel COVID-19 vaccines which will be effective for the newly discovered omicron variant and possibly more variants in the future in order to keep this disastrous virus under control.







